The objective was clear. We needed to speep up our iteration cycles. From raw data and labels to the prediction outputs that will help to train and improve our AI model.
Cleaning the messy data organisation and going to the cloud
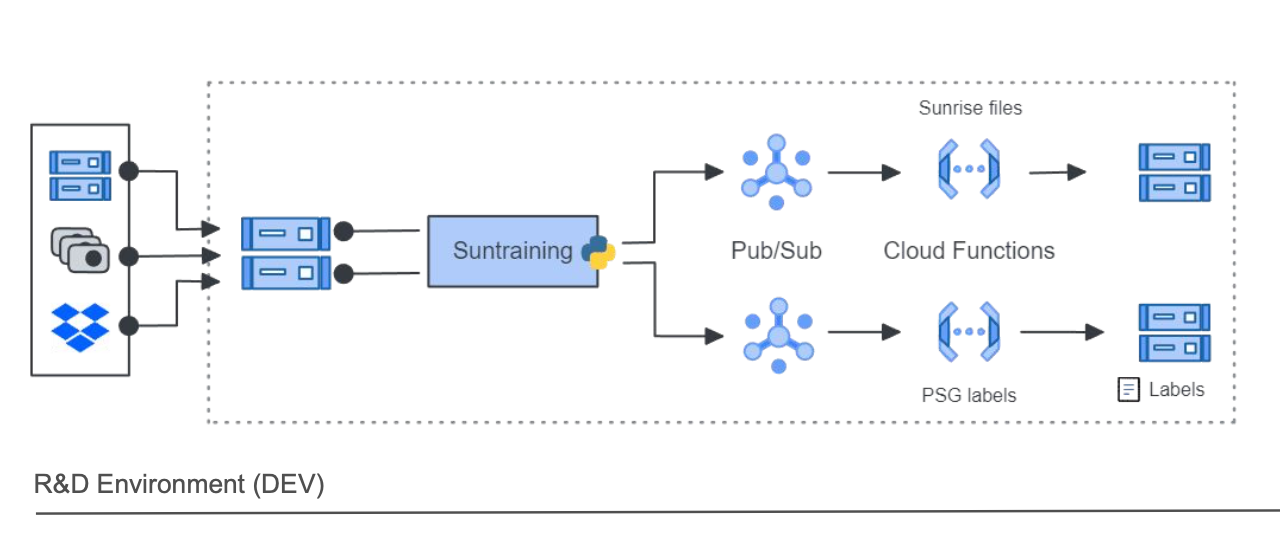
First, we cleaned up our data management. We decided to centralize in our data lake all the data spread over different buckets, dropox folders and hard drive in one bucket: the training bucket. This was our single source of truth, our Bronze layer (if you are familiar with medallion architecture from databricks). Obviously we brought some structure to it.
Next we moved from local computers to Google Cloud. To parallelize the pre-processing of our data, We used a service called Cloud Functions. It a serveless platform that package your code and spin up instances. You can scale up until 5000 thousands cloud functions in parallel and their processing times is 9 minutes maximum.
So we wrote the code necessary for the processing and packaged it to be shipped on Cloud Functions. Our approach was tool based. You will see that we developed a lot of CLI applications. The first CLI was leveraging Google Cloud python SDK to scrapes our data in the bucket, find the nights recorded both with the sunrise and polysomnography, anonymised the file, collect all metadata (encapsulated in our SunriseFilePayload, see below) and send all these informations to subscription topic defined in Pub/Sub.
PubSub can then push the metadata payload as an HTTP request to trigger our cloud function.
A snippet of the CLI code can be found below.
1 | def send_nights_to_pubsub( |
Anatomy of a cloud function
With this approach, we could now trigger thousands of cloud functions in parallel. With each function receiveing the metadata for one file, we could process thousands of files in a matter of minutes. Two Cloud Functions are used at this stage: one to process specifically the sunrise data, and one to process the polysomnography data.
Here is how we organize our code in our repository:
1 | . |
To deploy a cloud function you need to provide a main.py file with the code logic, a requirements.txt and if needed a .gcloudignore file to exclude any artifacts from the deployment in the cloud. The triggers.py contains the code shown just before. cf_cli.py was part our app.py, a CLI we developed using Typer. With this library you can decompose your cli application in several small pieces and unify the whole application in one file.
Typer is pretty neat to use and very easy to learn the API.
Our CLI appications in app.py contains commands to:
Deploy the cloud functions, which basically calls this
bashscript:1
2
3
4
5
6
7
8
9
10
11
SOURCE_DIR="cloud_functions/labels"
gcloud config set project sensav2;
gcloud functions deploy write-psg-labels-domino \
--region=europe-west1 \
--runtime=python310 \
--memory=2048 \
--source=${SOURCE_DIR} \
--entry-point=write_labels_for_domino \
--trigger-topic=write-labels-domino \
--timeout=540Send the metadata to the right PubSub topic so that we trigger the right Cloud Function processing.
The pre-processed sunrise and polysomnography data were then saved in two distinct buckets.
Predictions and features extration
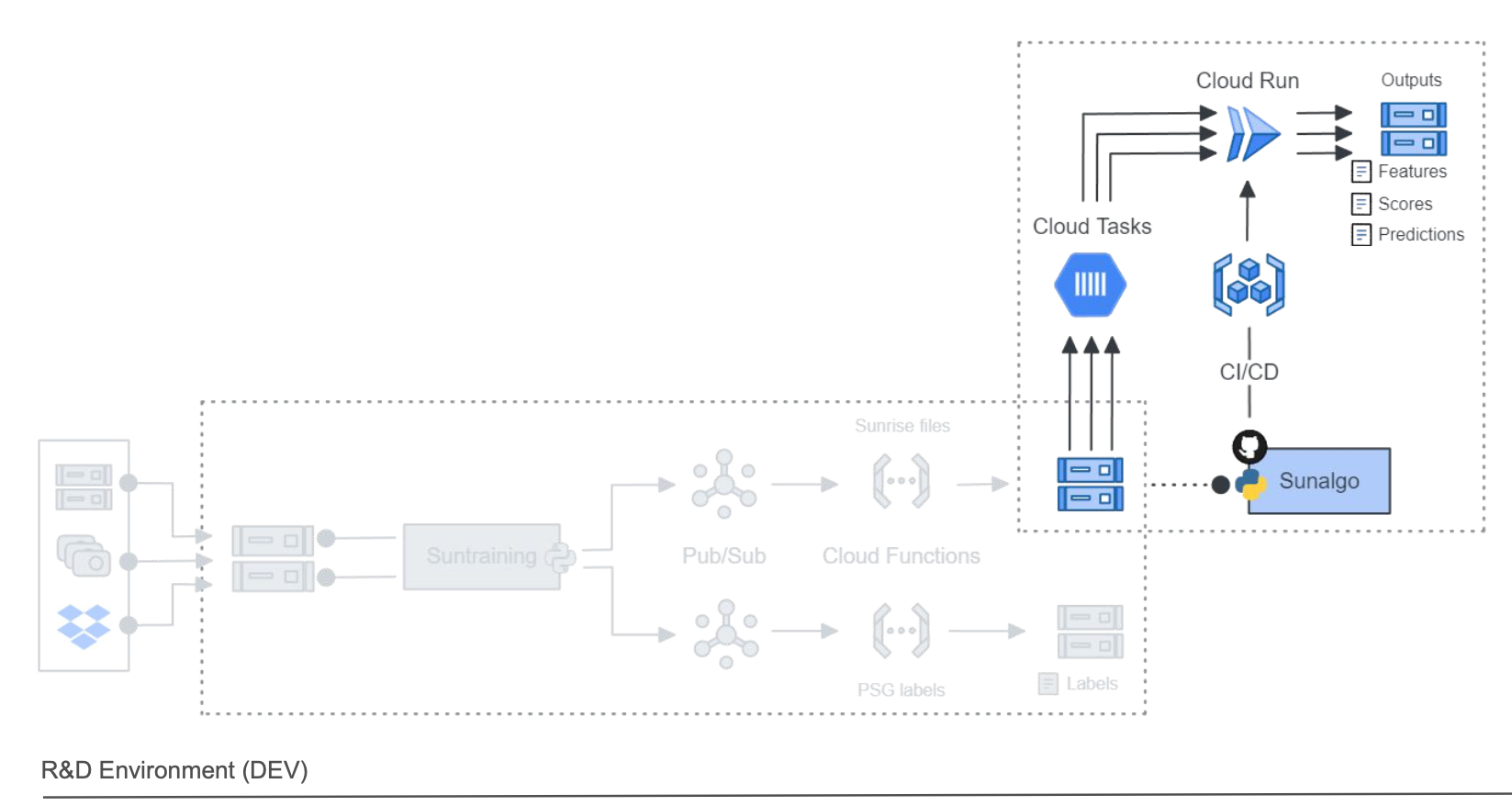
Once the sunrise data was pre-processed, it was ready to be used by our sunalgo application. The sunalgo was a monolithic service that accept the pre-processed sunrise data as input and generate the following outputs:
- Model features
- Model predictions
- Diverse clinical scores for sleep quality
Our sunalgo code was packaged as a docker container and stored in our artifact repository on Google Cloud. The container could then be used and deployed on Cloud Run instances. Cloud Run is a serveless service that can scale up and parallelize processing (up to 1000 instances), and scales down (even to zero) when no instance is needed. Our dockerfile looks like this:
1 | name: sunalgo-cicd-dev |
There is a lot to unpack here and the idea is not to enter on all the details. But you may pay attention to the way we (i) authetnicate with workload identity to let our github actions being able to interact with Google Cloud services, (ii) tag our docker image based on the
DevorProdenvironment and (iii) directly deploy on Cloud Run.
To not hit the limit of Cloud Run we decided to process our sunrise files by group of 800 tasks. To queue these tasks we used a service called Cloud Task. We could queue thousands tasks and cap it to a maximum of 800 active tasks. When a Cloud Run is finished it sends a message (HTTP payload) to the cloud task to receive the next task.
Cloud task is the type of service you can “click-click” to configure. Very easy and simple. No rocket science.
The trigger
With cloud task configured, our cloud run services up and running, we just needed to trigger the whole pipeline. And for that we used … a CLI application!
1 |
|
Below is a diagram of our whole pipeline.
Wrap up
By running just a couple of commands we could process more than 2000 files in some hours instead of days. The gain in time and efficiency was huge. At each step we tried to enforce good DevOps practices with continous integration and development using testing, containerization and Github Actions.
At this step we had another huge challenge to tackle: the training of the model. The generated predictions will be used with the labelled data and be ingested by our model for training. In the next article I will describe how we did that with Vertex.AI.